Large-scale histological image dataset toward stain and device-agnostic models

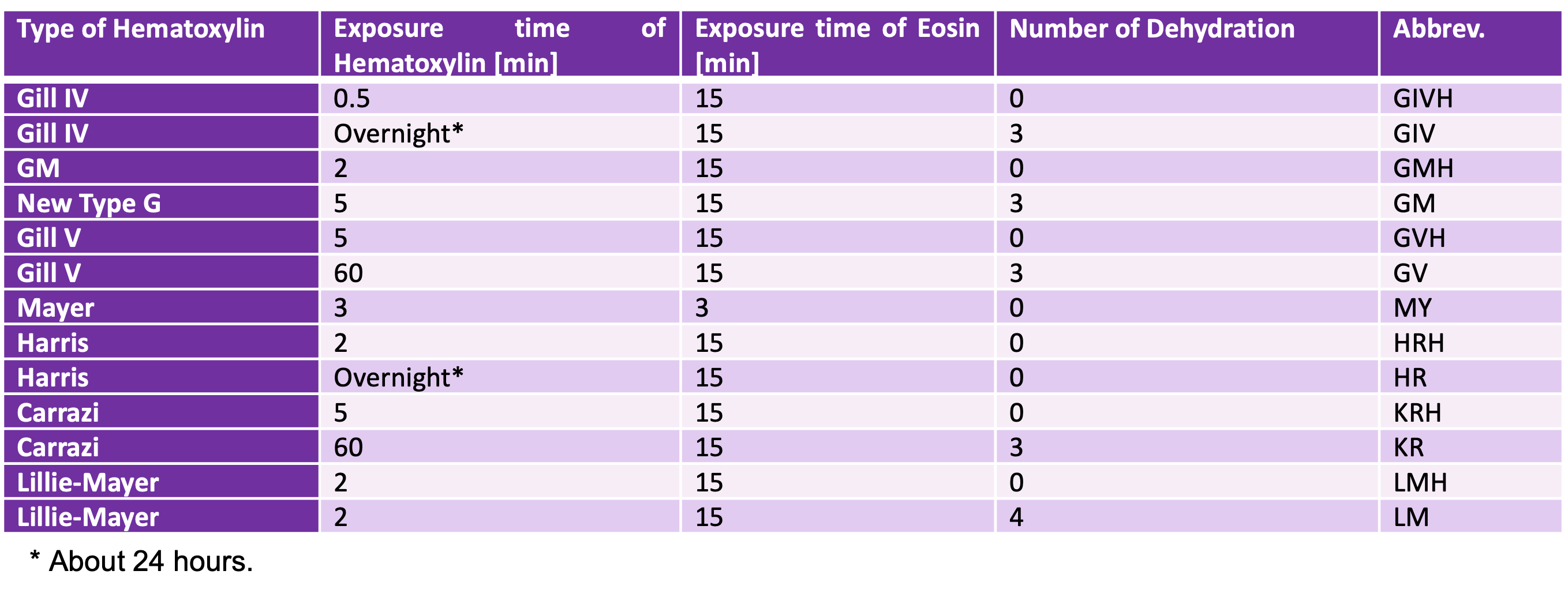
- Color and texture in digital pathology images are affected by H&E stain conditions (e.g. Harris or Carrazi) and digitalization devices (e.g. slide scanners or smartphones), which cause inter-institutional domain shifts.
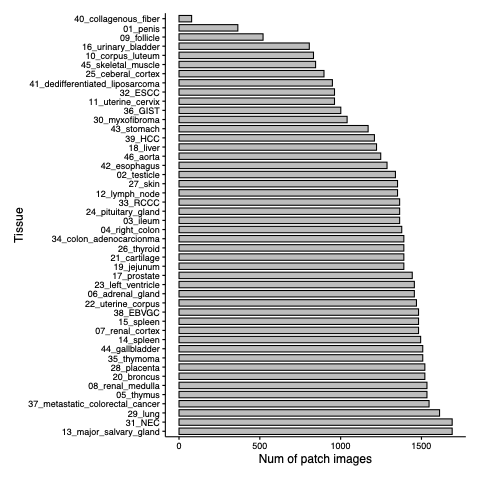
- PLISM is the first group-wised pathological image dataset that encompasses diverse tissue types stained under 13 H&E conditions, with multiple imaging media, including smartphones (7 scanners and 6 smartphones).
- PLSIM dataset was created for the evaluation of AI models’ robustness to domain shifts and development of robust AI models to them.
- Totally, the PLISM dataset is resulted in the creation of two subsets;
- 1. PLISM-sm, where smartphone images were used as queries to create image groups for each staining condition corresponding to each tile image.
- 2. PLISM-wsi, consisting of image groups for all staining conditions between WSIs for each tile image.
- Paper Link: Ochi, M., Komura, D., Onoyama, T. et al. Registered multi-device/staining histology image dataset for domain-agnostic machine learning models. Sci Data 11, 330 (2024). https://doi.org/10.1038/s41597-024-03122-5
- If you use the dataset for scientific work, please cite the above paper.
- We are pleased to share that Owkin has made the PLISM dataset tiles publicly available on Hugging Face.
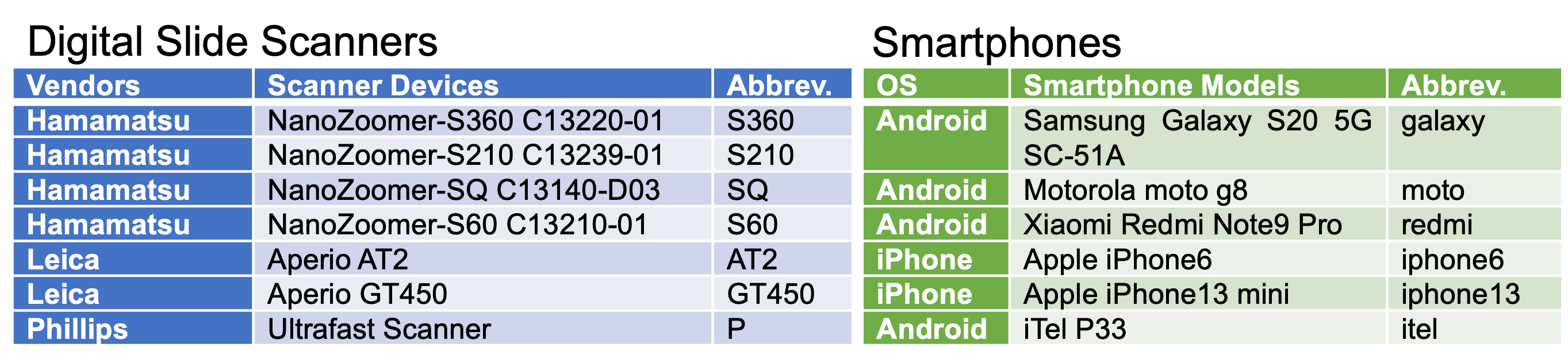
Sample tile images
Workflow & Tissue types
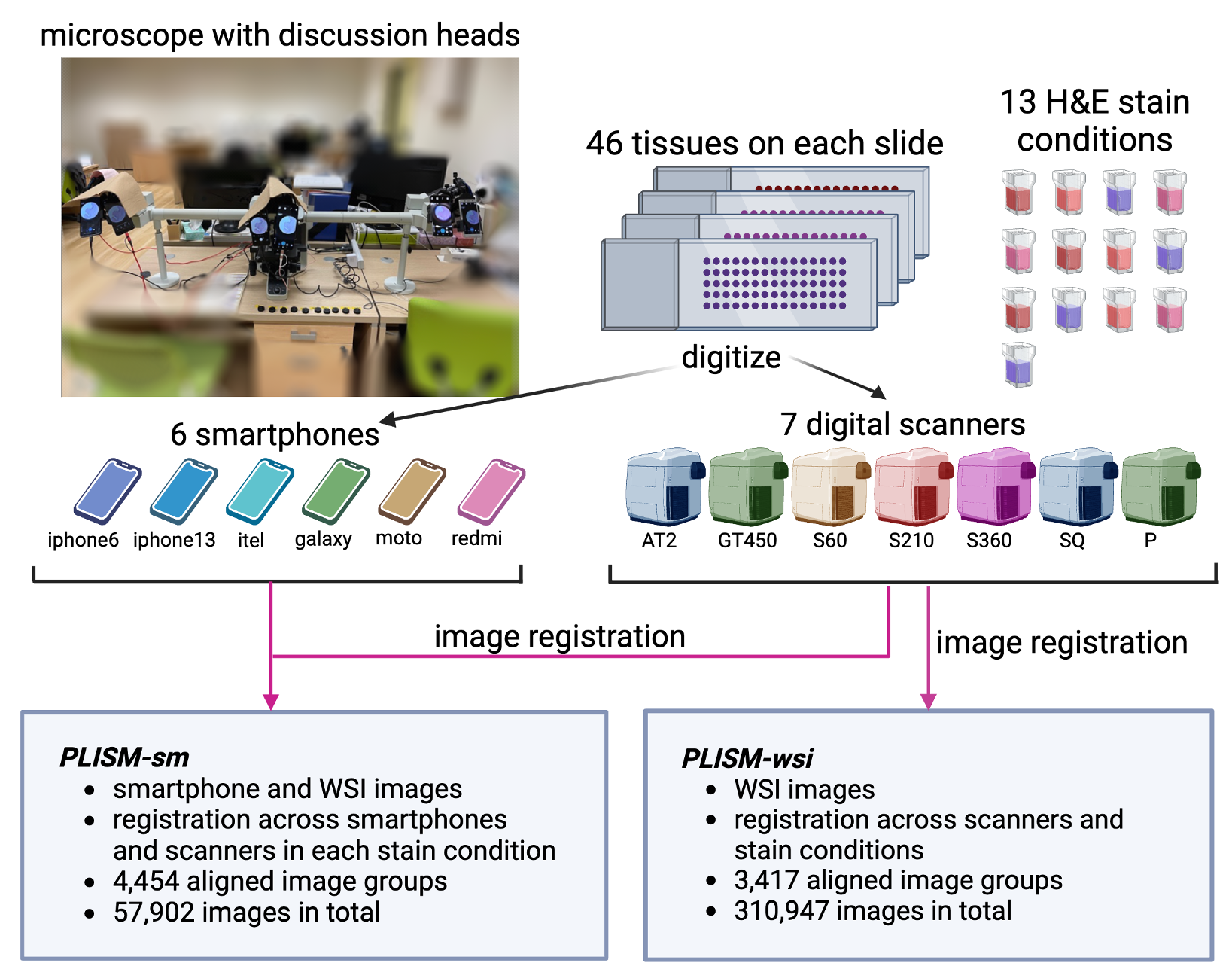
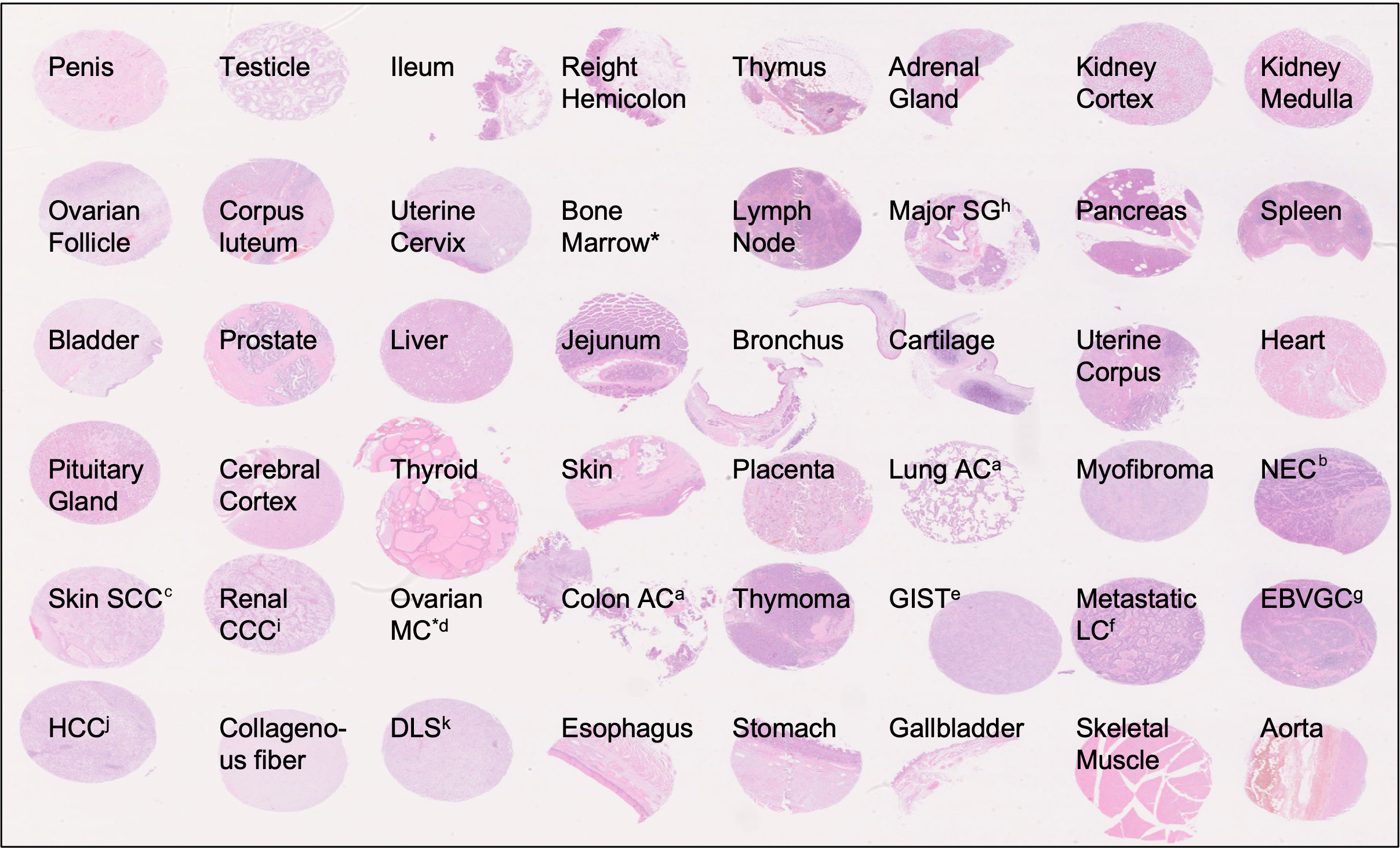
Breakdown of images by tissue